GenPharmaChain
Start Now


Revolutionizing Drug Discovery with AI-Driven Protein Engineering and Decentralized Computing


Stay healthy with Med+Med

01
Current issues in the pharmaceutical industry
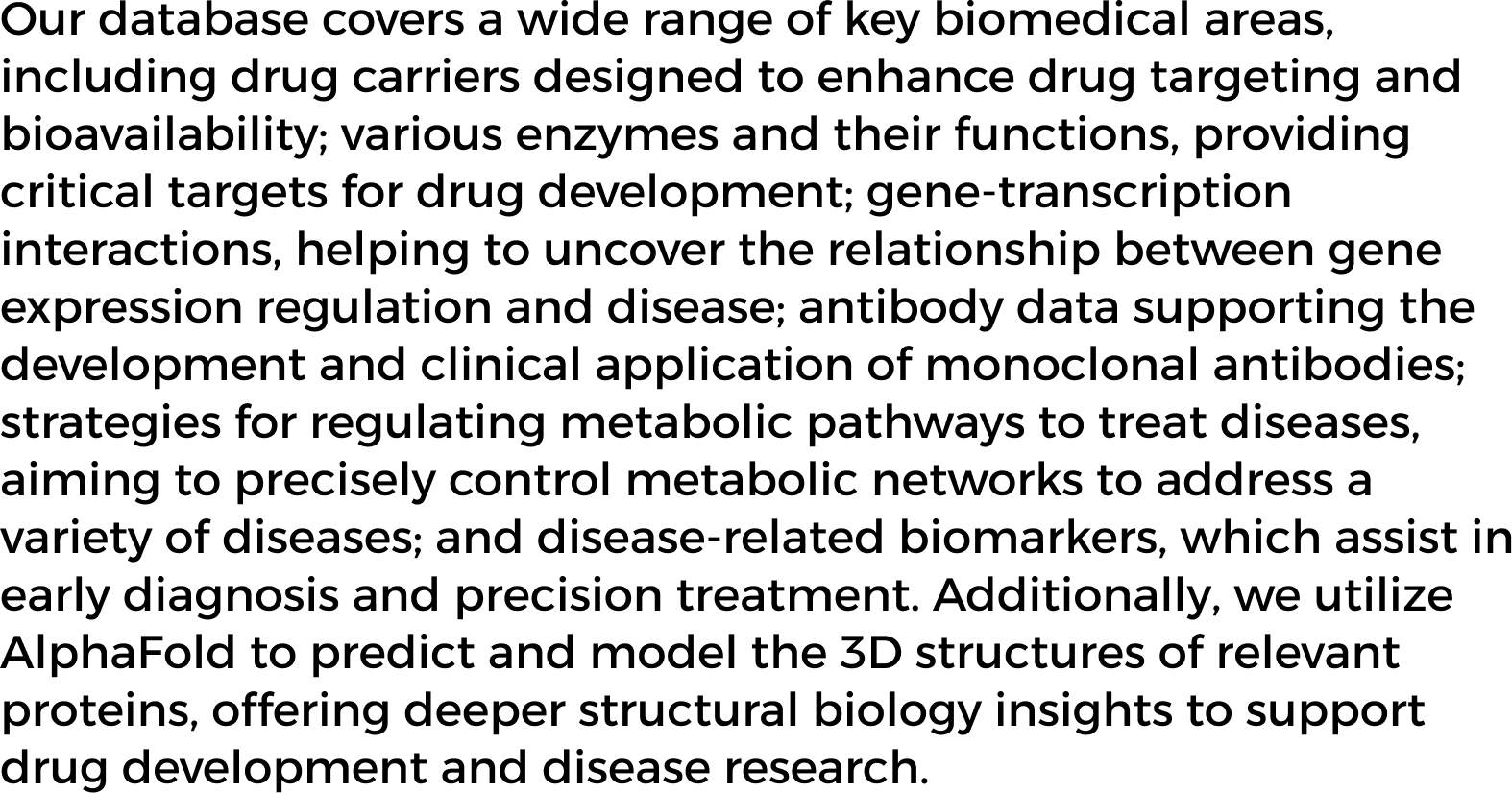
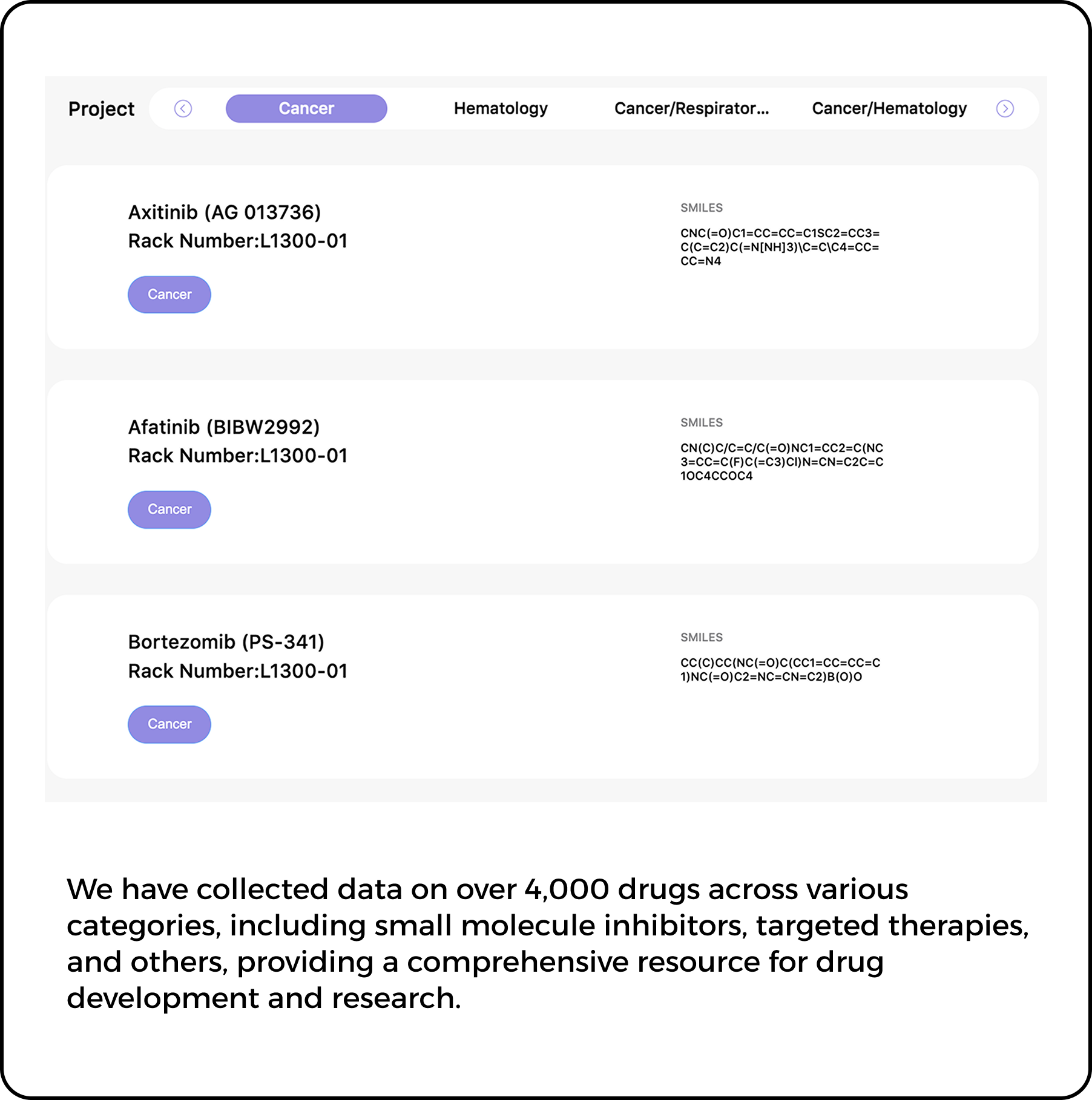
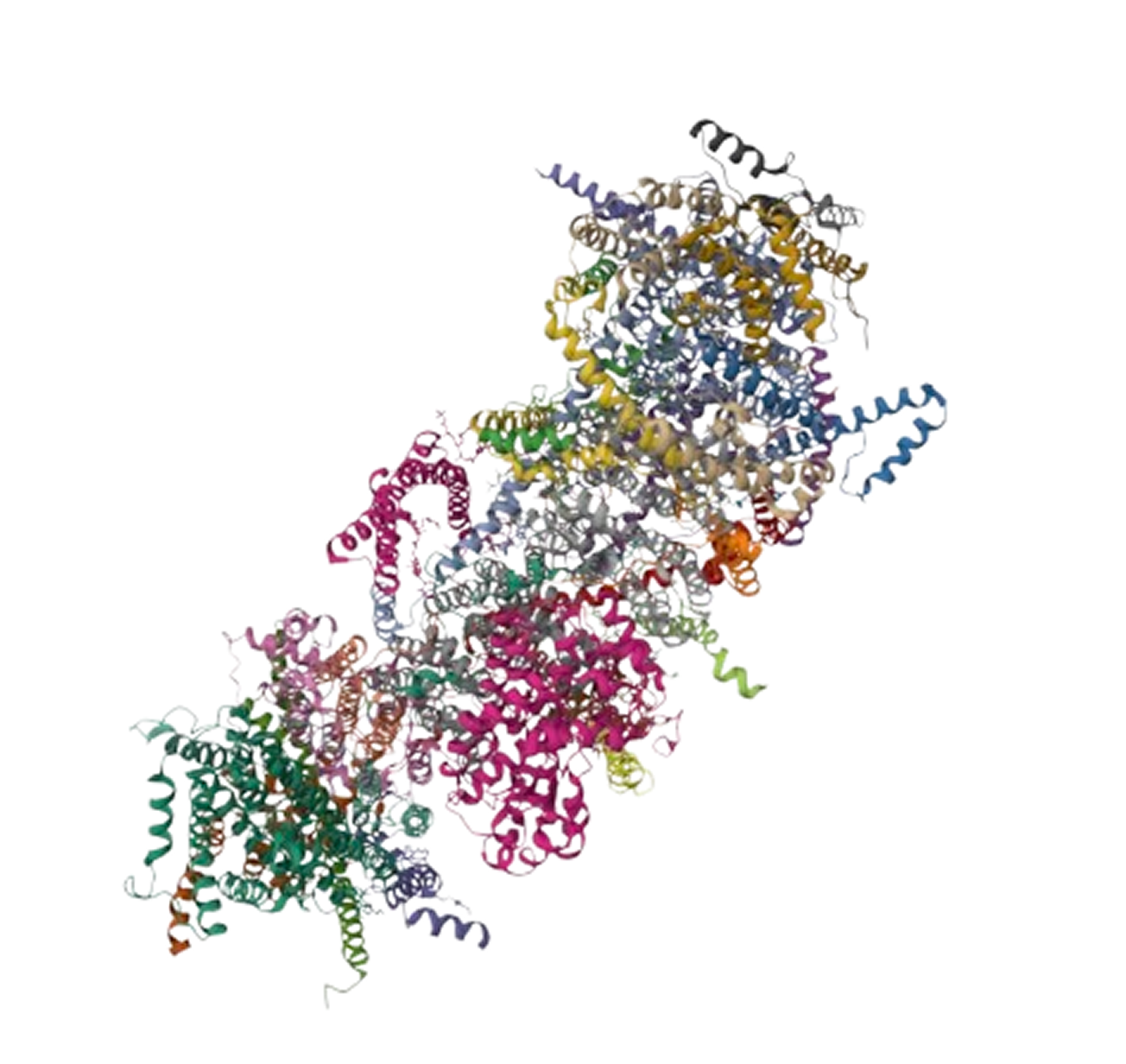
Protein prediction is crucial in modern medicine, particularly in precision medicine and drug development.
Proteins are key molecules that perform various functions in organisms, and protein prediction uses computer
modeling to predict their 3D structure and interactions, helping to understand their roles in cells and the
body. This process offers valuable insights into the causes of diseases.
Protein ® / drug develop / Trials
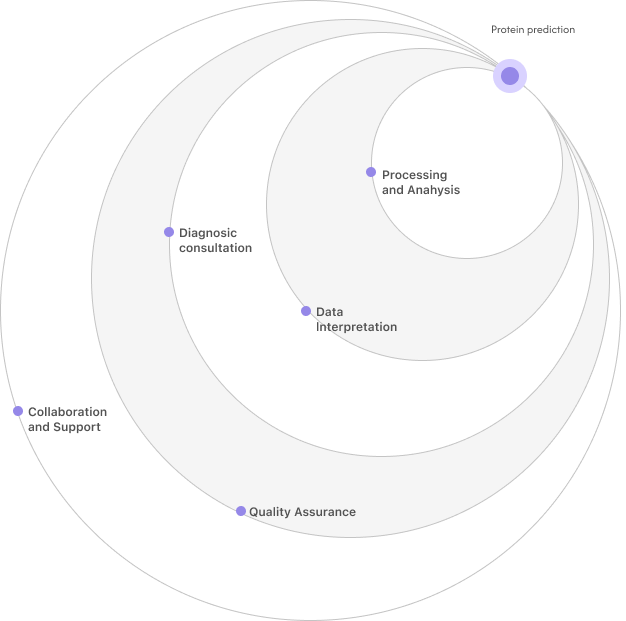
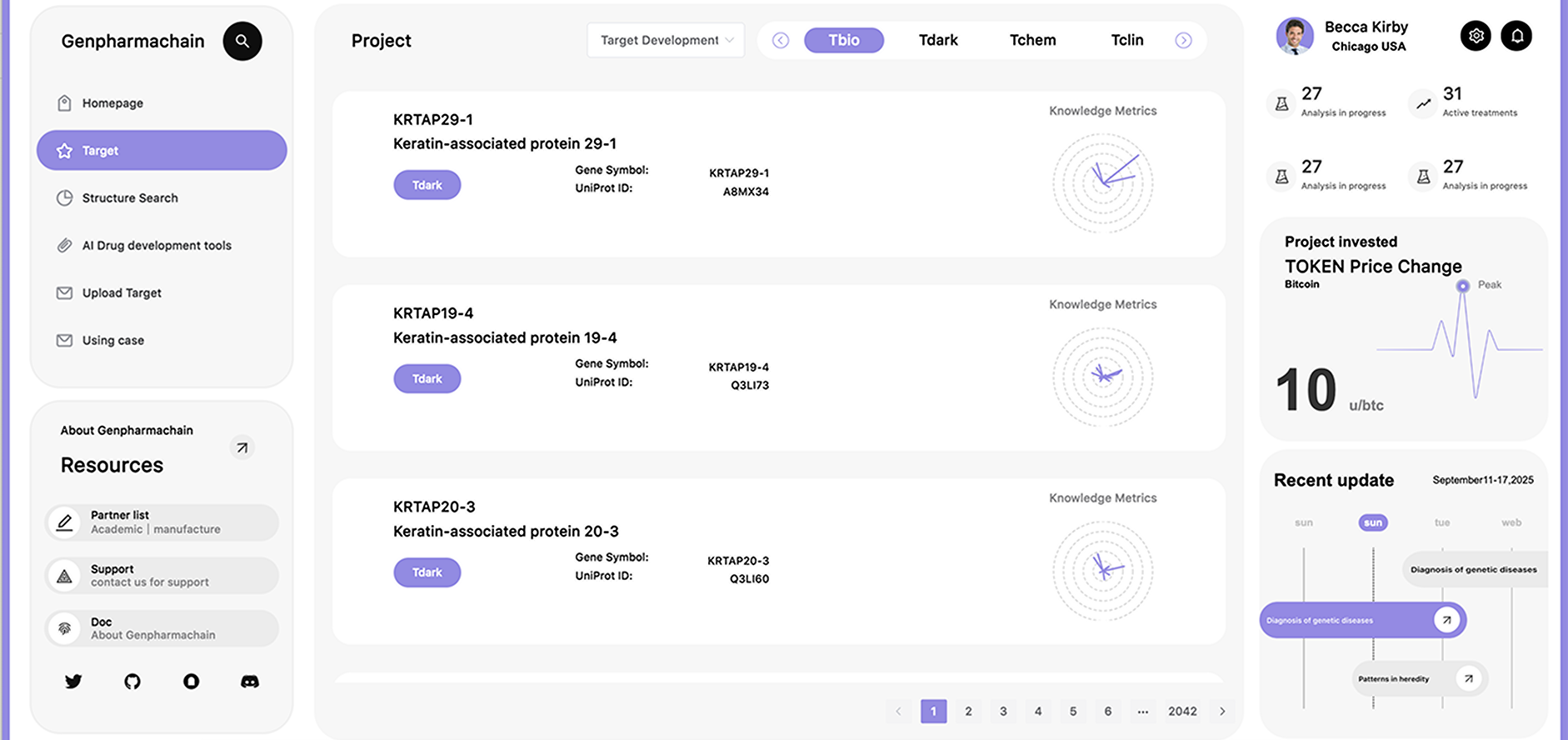
Our goal is to build a global decentralized platform where everyone can contribute their computational
resources to help with complex protein structure prediction and functional studies.
200M+
Database entire
47+
key organisms proteomes
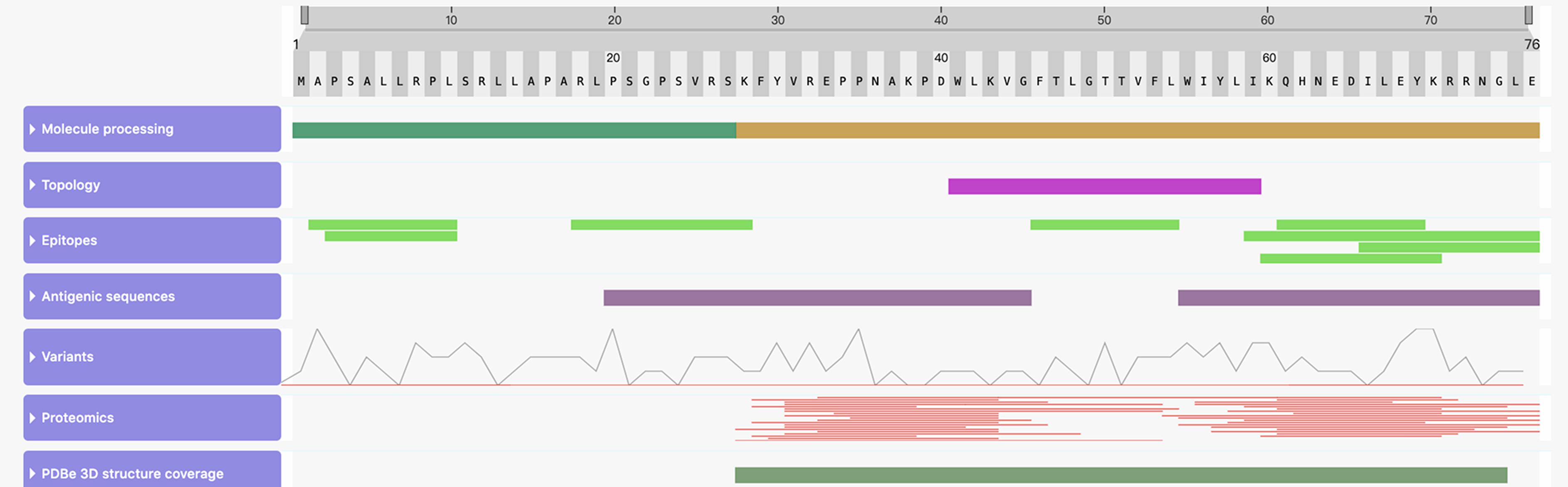
Reference:Cheng, J et al. Accurate proteome-wide missense variant effect prediction with
AlphaMissense. Science (2023).
Jumper, J et al. Highly accurate protein structure prediction with
AlphaFold. Nature (2021).Varadi, M et al. AlphaFold Protein Structure Database in 2024: providing structure
coverage for over 214 million protein sequences. Nucleic Acids Research (2024).
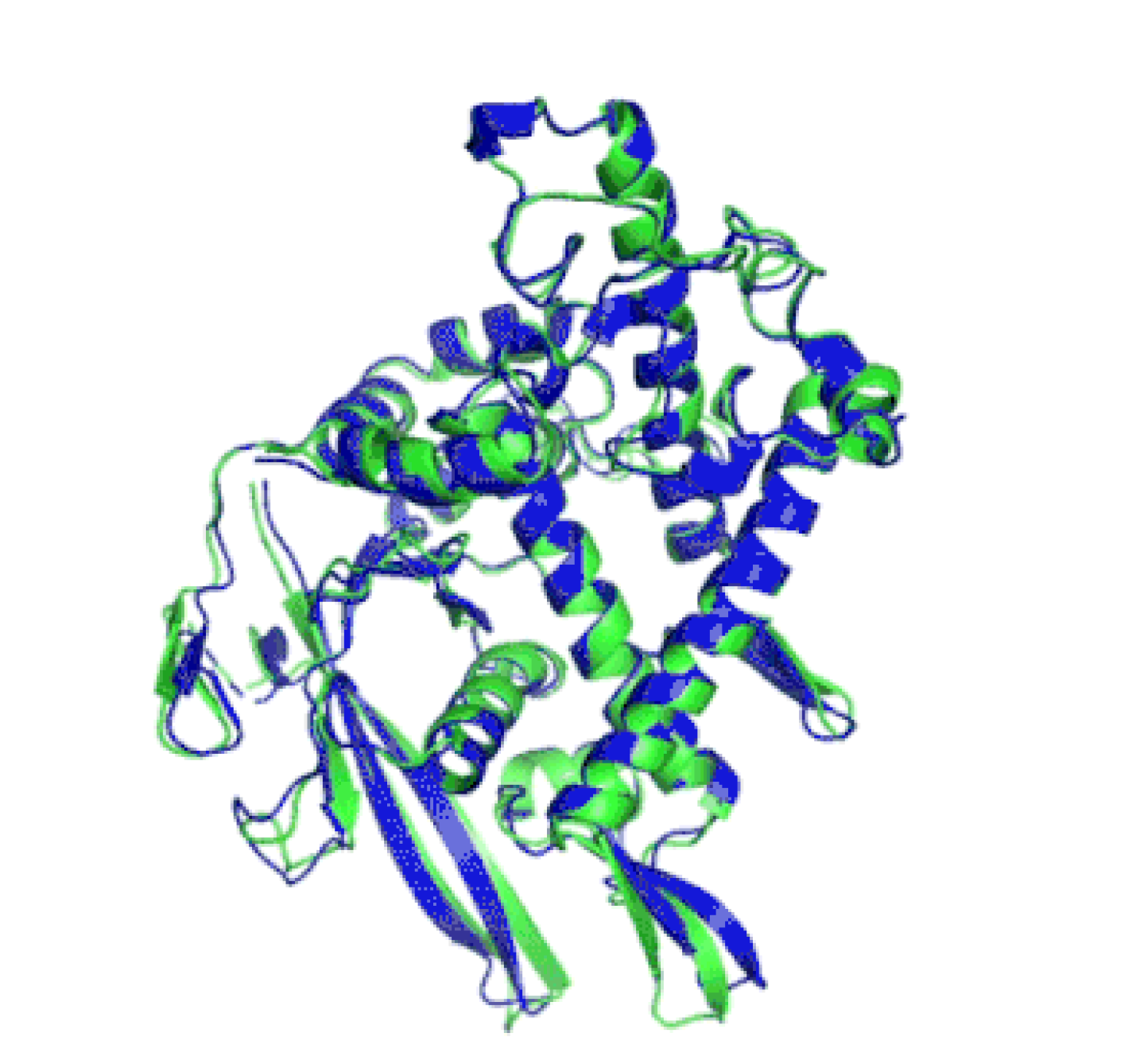
Protein 3D structure
Protein design
Start Now
Start Now
De novo design of protein topological structures
Applying Deep Learning Frameworks (such as AlphaFold, RoseTTAFold) for Large-Scale Fast
Modeling of Immunoglobulin Variable Region 3D Structures
Develop an automated pipeline capable of processing antibody, TCR, and nanobody sequence
datasets on the scale of 10510^5105, efficiently translating them into accurate three-dimensional structural
models.
GAN-based nanobody scaffold design platform to explore hypervariable region (HCDR) 3D
structures, engineer non-classical disulfide bonds, and optimize pH-dependent affinity maturation.
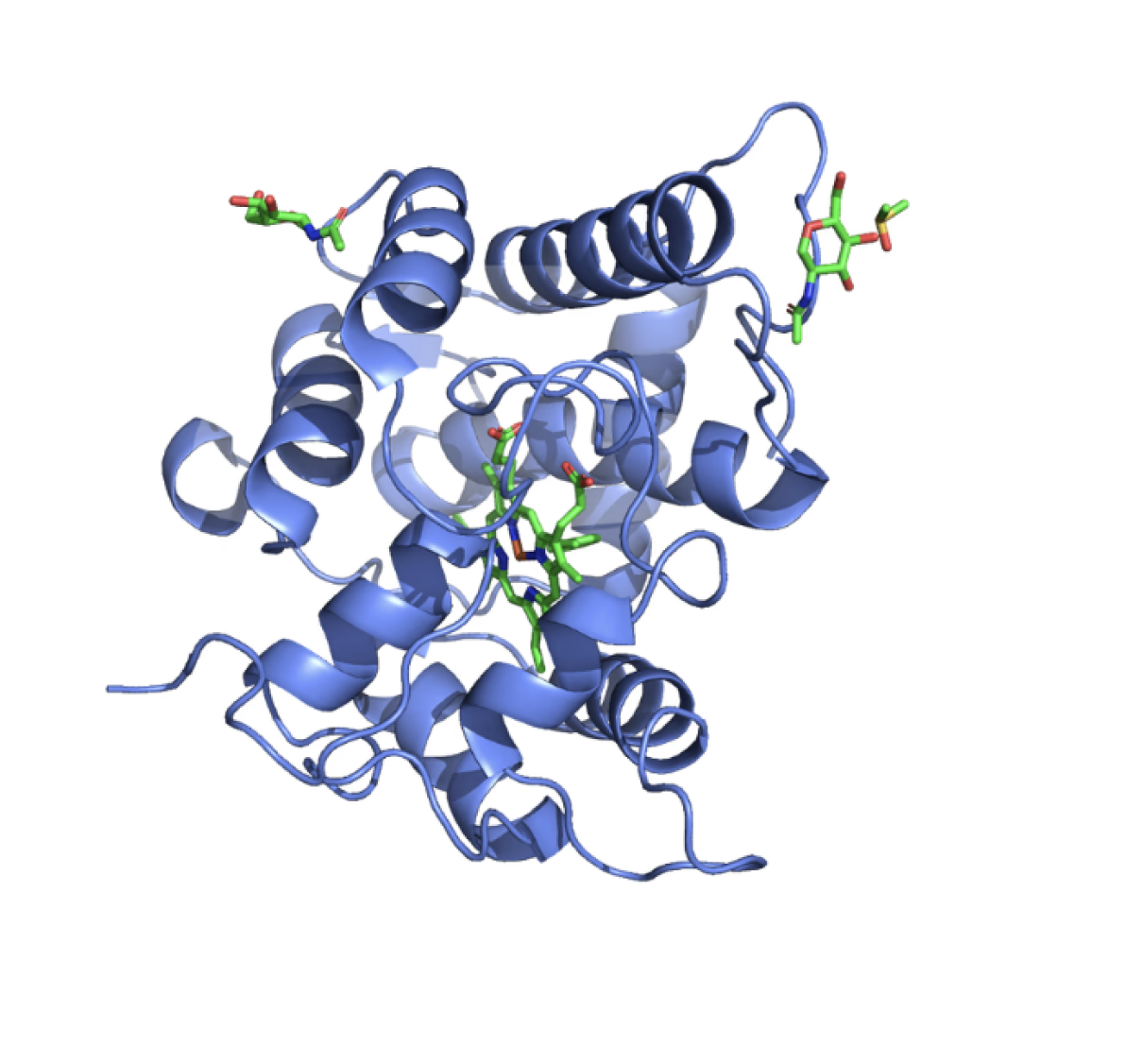
Protein topological structures
By generating new structures for motif scaffolds, it supports existing motifs on proteins.
The platform also incorporates partial diffusion, allowing for the diversification of part or all of the
protein to optimize various properties like solubility, thermal stability, and others.
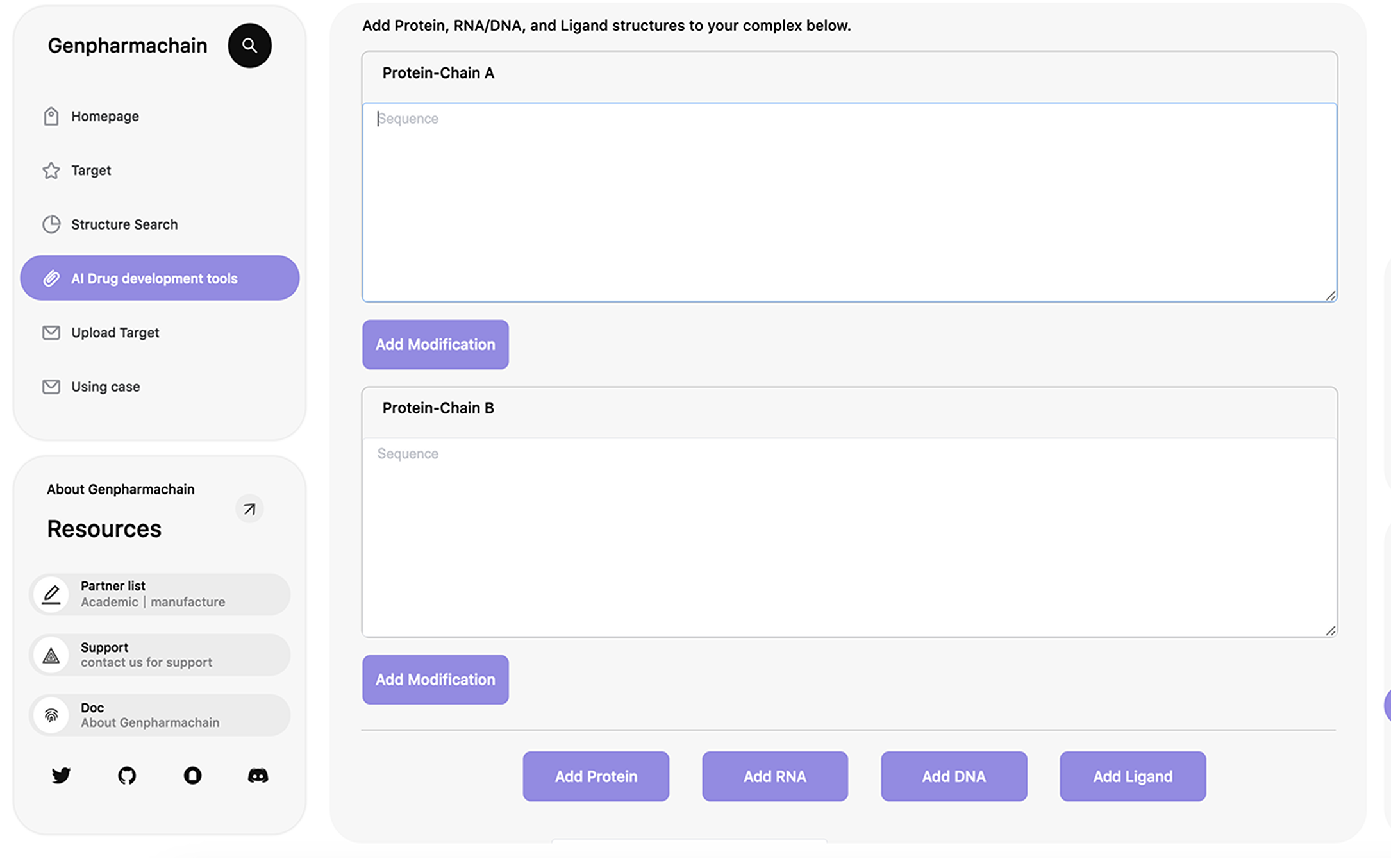
Generate sequences for a given protein scaffold, or upload multiple structures for batch
submission.
Choose the protein residues to be designed or fixed.
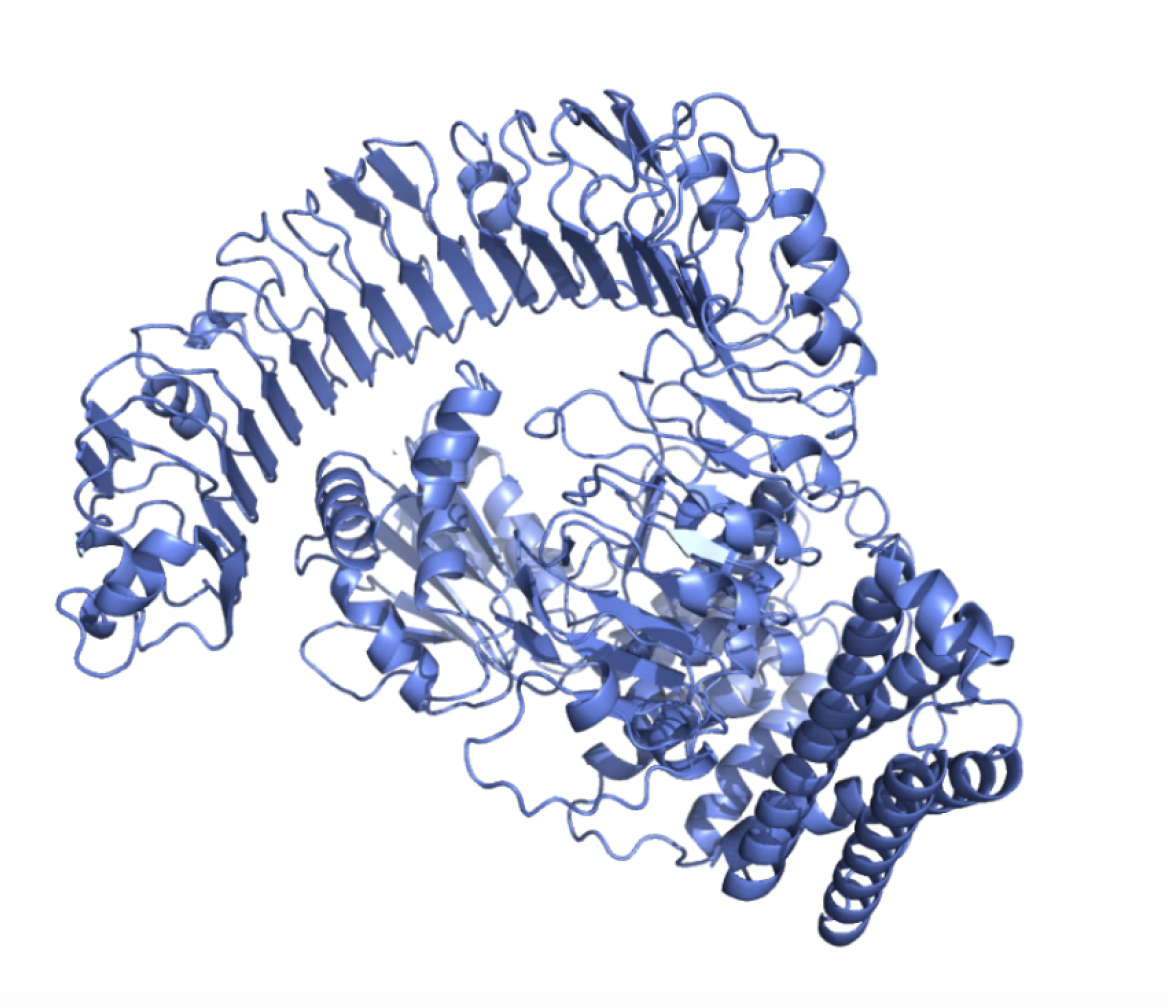
Protein topological structures
Predict how small molecules or candidate drugs bind to a known protein binding site, with
the site specified by the coordinates of the binding pocket.
Predict the binding affinity (kcal/mol) and the ligand pose.
Simulate protein-peptide systems to find optimal binding modes
Predict solubility, membrane permeability, and physicochemical characteristics
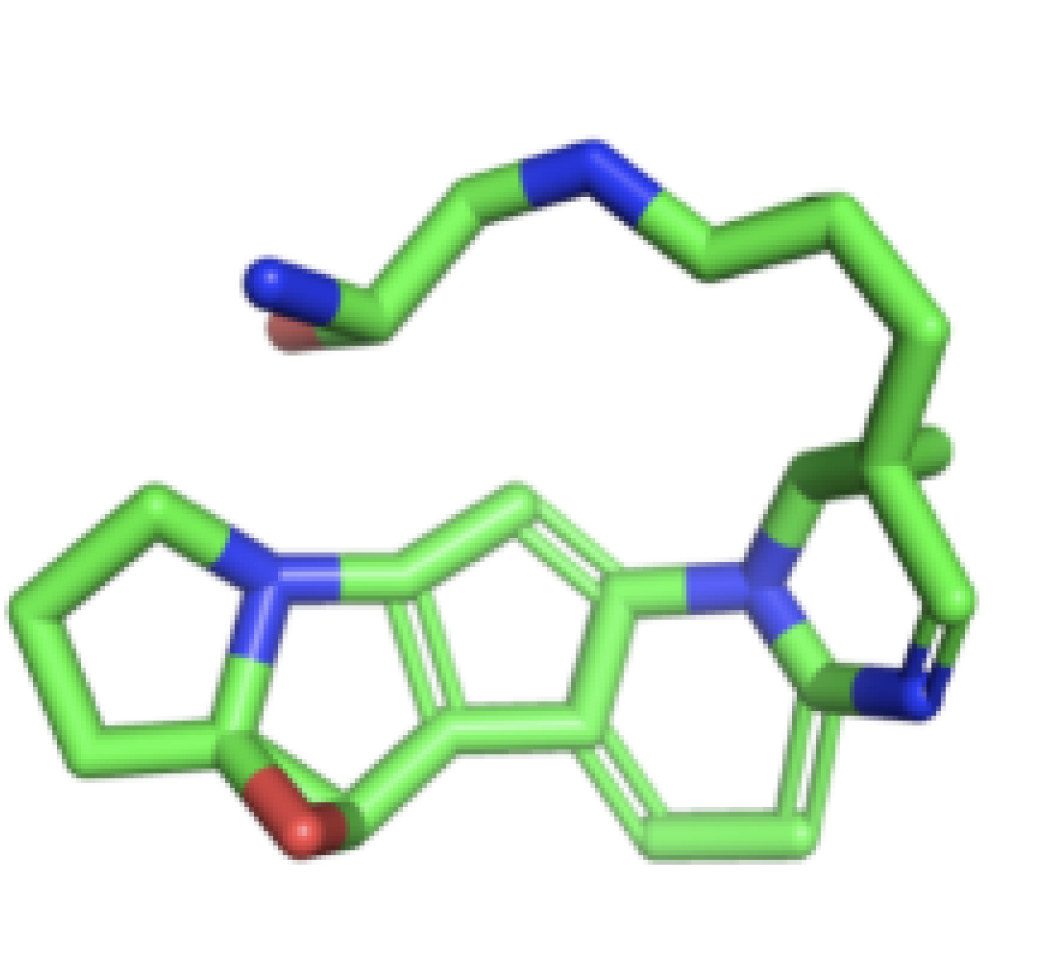
Drug molecular properties
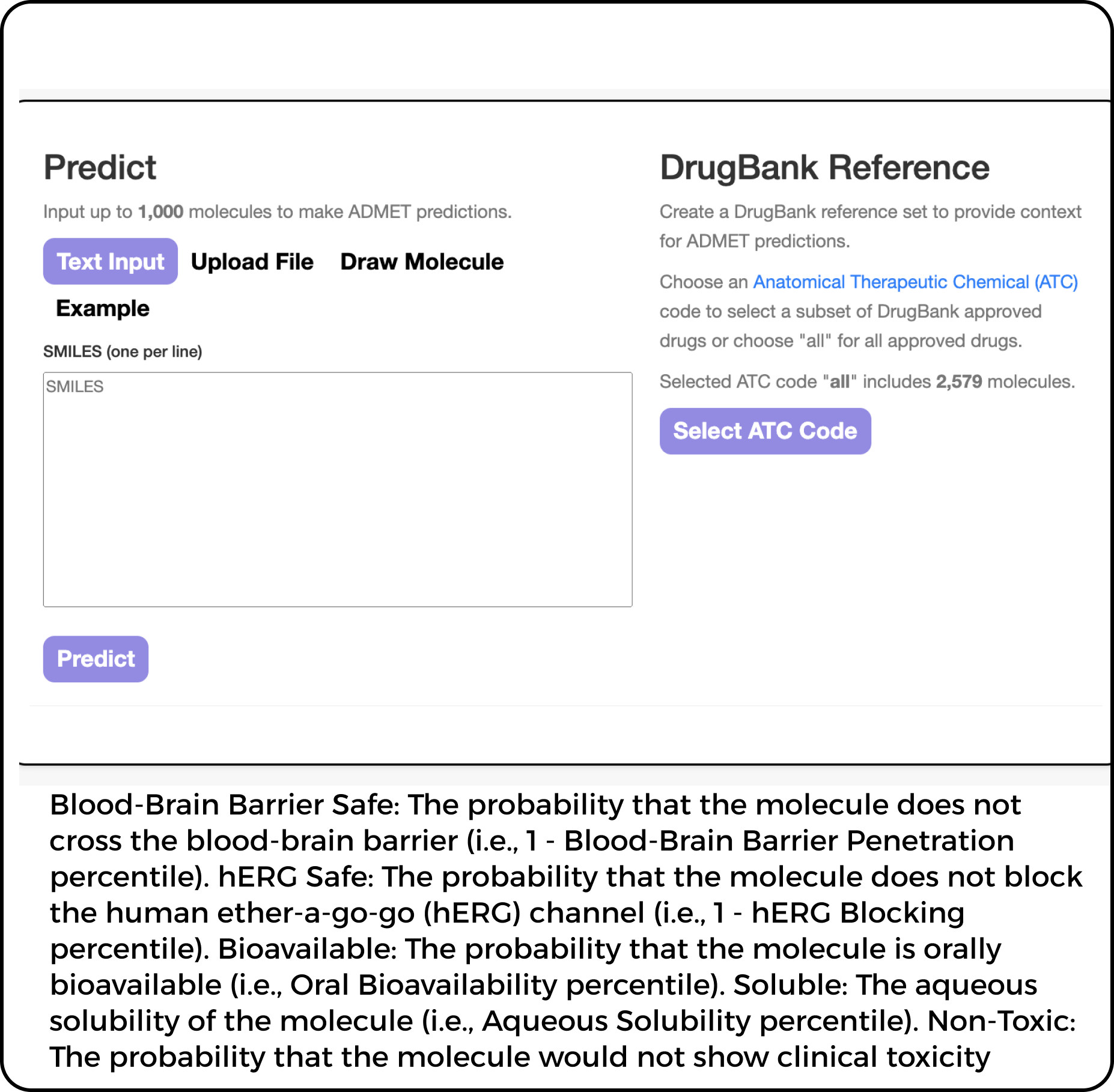
ADME represents the acronym for Absorption, Distribution, Metabolism, and Excretion - the
fundamental pharmacokinetic processes governing the dynamic disposition of drug compounds within biological
systems.
Absorption: Oral bioavailability prediction (Fa% ± 5% accuracy)
Distribution: Volume of distribution (Vd) modeling with PPB adjustments
Metabolism: CYP450 isoform profiling (IC50 determinations)
Excretion: Renal/hepatic clearance simulations
Protein research
 / PRECISION SERVICERE
/ PRECISION SERVICERE
 / Drug Production
/ Drug Production
 / PRECISION SERVICERE
/ PRECISION SERVICERE
 / Drug Production
/ Drug Production
Genpharamachain
Our Advantages



CONSITIT US